24 juli 2023: zie ook dit artikel: https://kanker-actueel.nl/circulerend-tumor-dna-ctdna-voorspelt-ziekteprogressie-bij-patienten-met-niet-kleincellige-longkanker-egfr-mutatie-die-worden-behandeld-met-tyrosinekinaseremmers.html
30 maart 2023: zie ook dit artikel: https://kanker-actueel.nl/circulerend-tumor-dna-ctdna-regelmatig-meten-en-combineren-met-een-weefselanalyse-biopt-geeft-beduidend-meer-informatie-over-veranderde-mutaties-bij-patienten-met-uitgezaaide-gevorderde-kleincellige-longkanker.html
24 juli 2023: Bron: Nature 21 november 2021
Meting van circulerend tumor-DNA (ctDNA) is een sterke voorspeller van al of niet een optreden van een recidief bij patiënten met niet-kleincellige longkanker. Wanneer het ctDNA een positieve uitslag geeft van overgebleven kankercellen dan is een verwacht recidief een heel stuk meer dan voor de patiënten die negatief scoorden in de ctDNA testen. ctDNA metingen werden gedaan direct na de operatie en gedurende de behandeling met chemotherapie en ook werden die ctDNA metingen gedaan bij patiënten die geen chemotherapie hadden gehad.
Dat blijkt uit een Chinese studie bij 103 operabele longkankerpatiënten stadium II/III waarbij gedurende een bepaalde periode regelmatig ctDNA is getest. Bij stadium II-III-patiënten heeft de postoperatieve ctDNA-positieve groep baat bij aanvullende chemotherapie als de ctDNA test positief was, terwijl ctDNA-negatieve patiënten een laag risico op een recidief hebben, ongeacht of er wel of geen chemotherapie is toegediend. Tijdens ziekte controle gaat ctDNA-positiviteit mediaan 88 dagen vooraf aan een radiologisch aantoonbaar recidief.
In onderstaande grafiek de verschillen grafisch weergegeven:
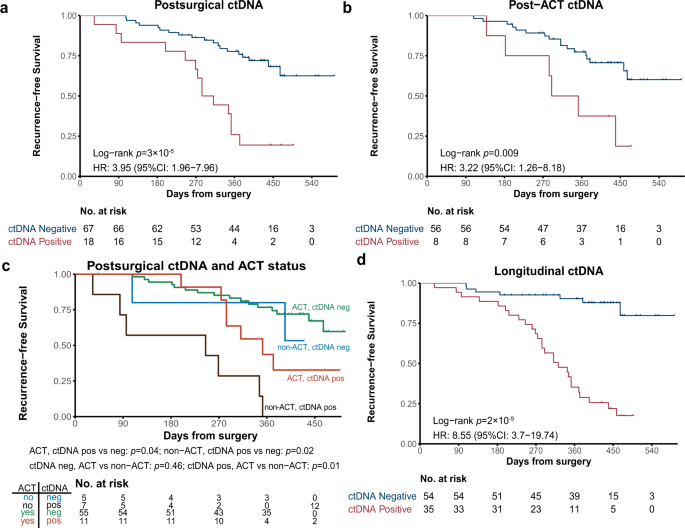
a Kaplan–Meier curve of recurrence-free survival (RFS) in patients stratified by postsurgical ctDNA status. p-value was calculated by the log-rank test. b Kaplan–Meier curve of RFS in patients stratified by post-ACT ctDNA status. p-value was calculated by the log-rank test. c Kaplan–Meier curve of RFS in stage II–III patients stratified by both ACT treatment and postsurgical ctDNA status. p-value was calculated by the log-rank test for each comparison without adjustments. d Kaplan–Meier curve of RFS in patients stratified by longitudinal ctDNA status. p-value was calculated by the log-rank test.
Het studieverslag, gepubliceerd in Nature geeft een gedetailleerd beeld van hoe is er getest en hoe de onderzoekers de verkregen gegevens hebben toegepast in deze studie. En hoe een ctDNA test van waarde kan zijn voor een vervolg behandeling na de operatie. Of juist geen behandeling om overbehandeling te voorkomen.
Het volledige studierapport is gratis in te zien. Klik op de titel voor het volledige studierapport:
- Article
- Open Access
- Published:
Dynamic recurrence risk and adjuvant chemotherapy benefit prediction by ctDNA in resected NSCLC
Nature Communications volume 12, Article number: 6770 (2021)
Abstract
Accurately evaluating minimal residual disease (MRD) could facilitate early intervention and personalized adjuvant therapies. Here, using ultradeep targeted next-generation sequencing (NGS), we evaluate the clinical utility of circulating tumor DNA (ctDNA) for dynamic recurrence risk and adjuvant chemotherapy (ACT) benefit prediction in resected non-small cell lung cancer (NSCLC). Both postsurgical and post-ACT ctDNA positivity are significantly associated with worse recurrence-free survival. In stage II-III patients, the postsurgical ctDNA positive group benefit from ACT, while ctDNA negative patients have a low risk of relapse regardless of whether or not ACT is administered. During disease surveillance, ctDNA positivity precedes radiological recurrence by a median of 88 days. Using joint modeling of longitudinal ctDNA analysis and time-to-recurrence, we accurately predict patients’ postsurgical 12-month and 15-month recurrence status. Our findings reveal longitudinal ctDNA analysis as a promising tool to detect MRD in NSCLC, and we show pioneering work of using postsurgical ctDNA status to guide ACT and applying joint modeling to dynamically predict recurrence risk, although the results need to be further confirmed in future studies.
Data availability
All raw targeted DNA-sequencing data have been deposited in the National Genomics Data Center (NGDC) under the accession code HRA001346. The deposited and publicly available data are compliant with the regulations of the Ministry of Science and Technology of the People’s Republic of China. The raw sequencing data contain information unique to individuals and are available under controlled access. Access to the data can be requested by completing the application form via GSA-Human System and is granted by the corresponding Data Access Committee. Additional guidance can be found at the GSA-Human System website [https://ngdc.cncb.ac.cn/gsa-human/document/GSA-Human_Request_Guide_for_Users_us.pdf]. Data used for survival analysis and joint model construction and evaluation are publicly available at https://github.com/cancer-oncogenomics/ctDNA-dynamic-prediction-lung-cancer. All specific mutation genomic locations and allele frequencies are available in Supplementary Data 2. Source data are provided with this paper.
Code availability
All analyses were performed using R version 4.0.2. R package survival (version 3.2-10) was used for survival analysis. R package JMbayes (version 0.8-85) was used for the construction and evaluation of joint models and cox models. Reference scripts to reproduce the results of this study is available at https://github.com/cancer-oncogenomics/ctDNA-dynamic-prediction-lung-cancer.
References
-
Bray, F. et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424 (2018).
-
Molina, J. R., Yang, P., Cassivi, S. D., Schild, S. E. & Adjei, A. A. Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clin. Proc. 83, 584–594 (2008).
-
Postmus, P. E. et al. Early and locally advanced non-small-cell lung cancer (NSCLC): ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 28, iv1–iv21 (2017).
-
NCCN Clinical Practice Guidelines in Oncology (version 8). National Comprehensive Cancer Network (2020).
-
Chao, M. & Gibbs, P. Caution is required before recommending routine carcinoembryonic antigen and imaging follow-up for patients with early-stage colon cancer. J. Clin. Oncol. 27, e279–e280 (2009).
-
Huang, K. et al. Radiographic changes after lung stereotactic ablative radiotherapy (SABR)–can we distinguish recurrence from fibrosis? A systematic review of the literature. Radiother. Oncol. 102, 335–342 (2012).
-
Group NM-aC. Adjuvant chemotherapy, with or without postoperative radiotherapy, in operable non-small-cell lung cancer: two meta-analyses of individual patient data. Lancet 375, 1267–1277 (2010).
-
Bradbury, P. et al. Postoperative adjuvant systemic therapy in completely resected non-small-cell lung cancer: a systematic review. Clin. Lung Cancer 18, 259–273 (2017).
-
Cortés, Á. A., Urquizu, L. C. & Cubero, J. H. Adjuvant chemotherapy in non-small cell lung cancer: state-of-the-art. Transl. Lung Cancer Res. 4, 191 (2015).
-
Garcia-Murillas, I. et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci. Transl. Med. 7, 302ra133 (2015).
-
Coombes, R. C. et al. Personalized detection of circulating tumor DNA antedates breast cancer metastatic recurrence. Clin. Cancer Res. 25, 4255–4263 (2019).
-
Reinert, T. et al. Analysis of plasma cell-free DNA by ultradeep sequencing in patients with stages I to III colorectal cancer. JAMA Oncol. 5, 1124–1131 (2019).
-
Yang, J. et al. Deep sequencing of circulating tumor DNA detects molecular residual disease and predicts recurrence in gastric cancer. Cell Death Dis. 11, 346 (2020).
-
Christensen, E. et al. Early detection of metastatic relapse and monitoring of therapeutic efficacy by ultra-deep sequencing of plasma cell-free DNA in patients with urothelial bladder carcinoma. J. Clin. Oncol. 37, 1547–1557 (2019).
-
Chaudhuri, A. A. et al. Early detection of molecular residual disease in localized lung cancer by circulating tumor DNA profiling. Cancer Disco. 7, 1394–1403 (2017).
-
Abbosh, C. et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 545, 446–451 (2017).
-
Hellmann, M. D. et al. Circulating tumor DNA analysis to assess risk of progression after long-term response to PD-(L) 1 blockade in NSCLC. Clin. Cancer Res. 26, 2849–2858 (2020).
-
Ibrahim, J. G., Chen, M.-H. & Sinha, D. Bayesian methods for joint modeling of longitudinal and survival data with applications to cancer vaccine trials. Stat. Sin. 14, 863–883 (2004).
-
Li, K., Furr-Stimming, E., Paulsen, J. S. & Luo, S., Group P-HIotHS. Dynamic prediction of motor diagnosis in Huntington’s disease using a joint modeling approach. J. Huntingt. Dis. 6, 127–137 (2017).
-
Ibrahim, J. G., Chu, H. & Chen, L. M. Basic concepts and methods for joint models of longitudinal and survival data. J. Clin. Oncol. 28, 2796–2801 (2010).
-
Asar, O., Ritchie, J., Kalra, P. A. & Diggle, P. J. Joint modelling of repeated measurement and time-to-event data: an introductory tutorial. Int. J. Epidemiol. 44, 334–344 (2015).
-
Zhang, X. C. et al. Comprehensive genomic and immunological characterization of Chinese non-small cell lung cancer patients. Nat. Commun. 10, 1772 (2019).
-
Chen, L. M., Ibrahim, J. G. & Chu, H. Sample size and power determination in joint modeling of longitudinal and survival data. Stat. Med. 30, 2295–2309 (2011).
-
Van Houwelingen, H. C. Dynamic prediction by landmarking in event history analysis. Scand. J. Stat. 34, 70–85 (2007).
-
Halabi, S., Li, C. & Luo, S. Developing and validating risk assessment models of clinical outcomes in modern oncology. JCO Precis Oncol. 3, https://doi.org/10.1200/PO.19.00068 (2019).
-
Rizopoulos, D., Molenberghs, G. & Lesaffre, E. M. Dynamic predictions with time‐dependent covariates in survival analysis using joint modeling and landmarking. Biometrical J. 59, 1261–1276 (2017).
-
Huang, Y., Li, W., Macheret, F., Gabriel, R. A. & Ohno-Machado, L. A tutorial on calibration measurements and calibration models for clinical prediction models. J. Am. Med. Inform. Assoc. 27, 621–633 (2020).
-
Newman, A. M. et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat. Med. 20, 548–554 (2014).
-
Abbosh. C. et al. Abstract CT023: Phylogenetic tracking and minimal residual disease detection using ctDNA in early-stage NSCLC: a lung TRACERx study. AACR (2020).
-
Chen, K. et al. Perioperative dynamic changes in circulating tumor DNA in patients with lung cancer (DYNAMIC). Clin. Cancer Res. 25, 7058–7067 (2019).
-
Bettegowda, C. et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 6, 224ra224–224ra224 (2014).
-
Razavi, P. et al. High-intensity sequencing reveals the sources of plasma circulating cell-free DNA variants. Nat. Med. 25, 1928–1937 (2019).
-
Xie, M. et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat. Med. 20, 1472–1478 (2014).
-
McKerrell, T. et al. Leukemia-associated somatic mutations drive distinct patterns of age-related clonal hemopoiesis. Cell Rep. 10, 1239–1245 (2015).
-
Genovese, G. et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N. Engl. J. Med. 371, 2477–2487 (2014).
-
Wang, Z. et al. Assessment of blood tumor mutational burden as a potential biomarker for immunotherapy in patients with non-small cell lung cancer with use of a next-generation sequencing cancer gene panel. JAMA Oncol. 5, 696–702 (2019).
-
Zhang, S. et al. Epidermal growth factor receptor (EGFR) T790M mutation identified in plasma indicates failure sites and predicts clinical prognosis in non-small cell lung cancer progression during first-generation tyrosine kinase inhibitor therapy: a prospective observational study. Cancer Commun. 38, 28 (2018).
-
Park, C. K., Cho, H. J., Choi, Y. D., Oh, I. J. & Kim, Y. C. A phase II trial of osimertinib in the second-line treatment of non-small cell lung cancer with the EGFR T790M mutation, detected from circulating tumor DNA: LiquidLung-O-Cohort 2. Cancer Res. Treat. 51, 777–787 (2019).
-
Taieb, J. et al. Prognostic value and relation with adjuvant treatment duration of ctDNA in stage III colon cancer: a post-hoc analysis of the PRODIGE-GERCOR IDEA-France trial. Clin. Cancer Res. 27, 5638–5646 (2021).
-
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
-
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
-
Koboldt, D. C. et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 22, 568–576 (2012).
-
Newman, A. M. et al. FACTERA: a practical method for the discovery of genomic rearrangements at breakpoint resolution. Bioinformatics 30, 3390–3393 (2014).
-
Rizopoulos, D. The R package JMbayes for fitting joint models for longitudinal and time-to-event data using MCMC. J. Stat. Software 72, https://doi.org/10.18637/jss.v072.i07 (2016).
Acknowledgements
We appreciate the support and participation of the physicians and patients in this study.
Author information
Authors and Affiliations
Contributions
B.Q. and S.G. conceived the study. B.Q., W.G., H.B., Y.S., F.T., Q.X. and S.G. provided project management and supervision. B.Q., W.G., F.Z., F.L., Y.J., Y.P. and F.T. provided or facilitated the accrual of patient samples, pathology, and/or clinical data. W.G., X.C. and H.B. performed bioinformatics and genomic analyses. B.Q., W.G., F.Z., F.L., Y.J. and Y.P. performed statistical analyses. W.G. and X.C. wrote the original draft, with input from all authors. B.Q., Y.X., H.B., S.G. and J.H. review and editing the manuscript.
Corresponding author
Ethics declarations
Competing interests
The following authors are employees of Nanjing Geneseeq Technology Inc. (Xiaoxi Chen, Hua Bao, Yang Xu, and Yang Shao). All remaining authors declare no competing interests.
Additional information
Peer review information Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
This article is cited by
-
Real world practice of postoperative radiotherapy for patients with completely resected pIIIA-N2 non-small cell lung cancer: a national survey of radiation oncologists in China
Radiation Oncology (2023)
-
Accuracy of minimal residual disease detection by circulating tumor DNA profiling in lung cancer: a meta-analysis
BMC Medicine (2023)
-
Individualized dynamic methylation-based analysis of cell-free DNA in postoperative monitoring of lung cancer
BMC Medicine (2023)
-
Practical recommendations for using ctDNA in clinical decision making
Nature (2023)
-
Integrative analysis of multi-omics data for liquid biopsy
British Journal of Cancer (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.
Gerelateerde artikelen
- ESMO 2024; aanbevolen abstracten gerelateerd aan longkanker door artsen, medisch specialisten en oncologen wereldwijd
- ASCO 2024: aanbevolen abstracten gerelateerd aan longkanker en asbestkanker (mesothelioma) door vooraanstaande oncologen wereldwijd
- Longkanker informatie algemeen
- Bestraling - radiotherapie bij longkanker: een overzicht van recente ontwikkelingen en belangrijke artikelen bij elkaar gezet. Update 15 april 2010
- Adagrasib met of zonder cetuximab geeft hoopvolle resultaten bij patiënten met darmkanker met gemuteerde KRAS G12C copy 1
- Angiogenese remmer, Vadimesam, ASA404, naast chemo, geeft geen enkel positief effect bij longkanker, aldus fase 3 studie resultaten
- Alimta - Pemetrexed - goedgekeurd door FDA en Europa voor niet-klein-cellige longkanker als onderhoudstherapie. Pointbreak fase 3 studie echter stopgezet wegens falen van Alimta
- Biomarker test op EGFR en ALK bij longkanker voor behandeling start is kosteneffectief in gewonnen levensjaren en kwaliteit van leven in vergelijking met eerst chemo en dan pas biomarkertest. copy 1
- Selpercatinib (LOXO-292), geeft alsnog uitstekende resultaten (68 procent objectieve respons) met langdurende remissies bij zwaar voorbehandelde RET fusie-positieve niet-kleincellige longkanker met ook hersenuitzaaiingen
- Chemo bij longkanker: overzicht van recente ontwikkelingen en belangrijke studies
- Chinese kruiden als aanvulling bij BAIC - chemo embolisatie bij bronchiale longkanker geeft significant betere overleving - verdubbeling van twee jaars overleving van 24 procent naar 51 procent, aldus gerandomiseerde studie
- ctDNA = circulerend tumor-DNA meet en voorspelt al of niet minimale overgebleven kankercellen bij patienten met niet-kleincellige longkanker en is belangrijk voor post operatieve behandeling
- Circulerend tumor-DNA (ctDNA) voorspelt ziekteprogressie bij patiënten met niet-kleincellige longkanker (EGFR - mutatie) die worden behandeld met tyrosinekinaseremmers
- COPD - Chronic Obstructive Pulmonary Disease: Roflumilast verbetert longfunctie bij COPD patiënten aldus fase III studie bij 1411 COPD patiënten
- Cryosurgery - bevriezen van tumoren - bij longtumoren ook succesvol en hoopgevend.
- Diagnose en oorzaak van longkanker: een overzicht van artikelen en recente ontwikkelingen. Scroll in linkerkolom voor artikelen
- Durvalumab + chemotherapie voor operatie gevolgd door alleen durvalumab ipv chemo geeft meer complete remissies bij patiënten met operabele niet-kleincellige longkanker
- EGFR remmers zoals Tarceva - Erlotinib en Iressa - Gefitinib: een overzicht van artikelen en recente ontwikkelingen.
- Epigenetische behandeling met Azacitidine en entinostat zorgt voor spectaculair goede resultaten bij uitbehandelde patienten met niet-klein-cellige longkanker
- Genetisch onderzoek: Next Generation Sequencing (NGS) moet longkankerpatienten betere en gerichtere behandeling geven copy 1
- Hyperthermie plus voedingsprogramma (kytogeen dieet) plus hyperbare zuurstoftherapie aanvullend op chemo geeft uitstekende resultaten op mediane overall overleving (42 maanden) bij patienten met gevorderde niet-kleincellige longkanker copy 1 copy 1
- Hormonen: Er is een verband tussen longkanker bij vrouwen en hormonale factoren. Na 5 jaar bleken vrouwen uit de hormoongroep een grotere kans op overlijden te hebben dan vrouwen die geen hormonen gebruikten. 67 vs 39 doden van de longkankerpatienten.
- Immuuntherapie en vaccinaties bij longkanker: een overzicht van belangrijke studies en artikelen en recente ontwikkelingen.
- LITT - Laser-induced Interstitial Thermotherapy lijkt uitstekende aanpak voor longkanker met gebruikmaking van uiterst fijne apparatuur en waarbij de kanker tumor voor tumor wordt aangepakt. Dus niet alles tegelijk.
- Operatie: Longkankerpatienten met slecht 1 uitzaaiing, zouden daaraan toch geopereerd moeten worden en zijn genezend te behandelen, blijkt uit nieuwe meta analyse
- Palliatieve zorg plus oncologische zorg vanaf diagnose bij longkankerpatienten leidt tot langere overleving, betere kwaliteit van leven, betere voedingstatus en betere psychosociale gesteldheid in vergelijking met alleen standaard oncologische zorg
- PDT - Fotodynamische Therapie kan zeker ook bij longkanker een succesvolle behandeling zijn
- Repotrectinib, een TRK remmer, geeft uitstekende resultaten bij patiënten met gevorderde niet-kleincellige longkanker (Respons van 93 procent) en patienten met solide tumoren met een ROS1-fusie copy 1
- R.F.A. - Radio Frequency Ablation - een operatietechniek waarbij tumoren worden vernietigd door hitte via holle naalden is ook met succes toe te passen bij longkankertumoren.
- Tremelimumab (Imjudo) plus durvalumab (Imfinzi) naast op platina gebaseerde chemotherapie verbetert ziekte progressievrije tijd en overall overleving bij patienten met vergevorderde longkanker en krijgt goedkeuring van FDA.
- Tumor Treating Fields aanvullend op standaard behandeling geeft langere overleving in vergeliking met alleen standaard behandeling bij patienten met niet-kleincellige longkanker, bewijst fase 3 studie LUNAR
- Tyrosine Kinase remmers (TKI) bij longkanker met EGFR mutaties. Een overzicht van artikelen van verschillende TKI remmers.
- Zometa - Zoledronic Acid geeft betere resultaten dan APD - Pamidronate bij botproblemen (breuken bv.), langere tijd tot eerste botuitzaaiingen zich voordoen en vermindert significant de botpijn bij in bot uitgezaaide longkanker
- Regulier - Longkanker: overzicht van recente ontwikkelingen en belangrijke studies en artikelen binnen reguliere oncologie voor longkanker.



Plaats een reactie ...
Reageer op "ctDNA = circulerend tumor-DNA meet en voorspelt al of niet minimale overgebleven kankercellen bij patienten met niet-kleincellige longkanker en is belangrijk voor post operatieve behandeling"