Mocht u kanker-actueel de moeite waard vinden en ons willen ondersteunen om kanker-actueel online te houden dan kunt u ons machtigen voor een periodieke donatie via donaties: https://kanker-actueel.nl/NL/donaties.html of doneer al of niet anoniem op - rekeningnummer NL79 RABO 0372931138 t.n.v. Stichting Gezondheid Actueel in Amersfoort. Onze IBANcode is NL79 RABO 0372 9311 38
Elk bedrag is welkom. En we zijn een ANBI instelling dus uw donatie of gift is in principe aftrekbaar voor de belasting.
22 februari 2022: lees ook dit artikel: https://kanker-actueel.nl/NL/alzheimerpatienten-missen-goede-bacterie-in-hun-darmen-die-gezonde-mensen-wel-hebben-veel-vezels-en-gevarieerd-eten-zou-alzheimer-kunnen-voorkomen.html
8 september 2017: Lees ook dit artikel:
7 januari 2016: Bron: Front Microbiol. 2015; 6: 1050. Published online 2015 Oct 6. doi: 10.3389/fmicb.2015.01050
Veel mensen weten wel dat je darmflora - bacteriën in de darm - een grote rol spelen in onze gezondheid. En veel mensen weten ook wel dat gevarieerd eten gezonder is dan elke dag hetzelfde. Zelfs als dat allemaal gezond en verantwoord eten is. Bekendste euvel van vegetariërs is bv. dat ze bepaalde eiwitten, bepaalde vitamines of mineralen of bv. ijzer uit vlees tekort komen als ze dat niet compenseren met specifieke voedingsproducten zoals noten, bepaald fruit enz. Ik ben geen voedingsdeskundige maar op internet is heel veel daarover te vinden of vraag uw huisarts of behandelend arts of diëtisten in uw omgeving.
Nog niet zo lang geleden lag de nadruk op eten met voldoende zogeheten macronutriënten zoals koolhydraten, eiwitten en vetten en op de zogeheten micronutriënten zoals vitamines, mineralen, spoorelementen en vezels. Daarom was gevarieerd eten volgens de schijf van vijf zo belangrijk om uit elk voedingsproduct weer iets anders te halen. De laatste jaren wordt door deskundigen bv. geadviseerd om zo weinig mogelijk koolhydraten te eten en minder vlees en meer groenten en fruit. De schijf van vijf is daarop ook recent beetje aangepast, met name minder vlees eten. Een goeie zaak lijkt mij gezien de mindere kanten van vlees eten dat kankerverhogend werkt.
Toch lijkt het ook raadzaam om niet zomaar over te gaan op een eenzijdig voedselpatroon.
Hoe rijker je darmflora, hoe gezonder je spijsvertering - metabolisme. Bv. Hilde Maris schrijft in haar boek: Kanker begrijpen - De wijsheid van het lichaam het volgende:
"De bacteriën in de darm voeden zich niet allemaal met dezelfde nutriënten. Sommige bacteriën voeden zich met koolhydraten of vezels, andere met eiwitten. Als je eenzijdig eet, krijgen bepaalde bacteriën te veel en andere te weinig voedsel. Dat betekent dat sommige bacteriën zich sterk kunnen vermenigvuldigen en andere afsterven. Verschillende bacteriën hebben verschillende taken, zoals het moduleren van de immuniteit, het remmen van inflammatie (ontstekingen), het controleren van de bloedsuikerspiegel, het reguleren van honger en verzadiging, de afbraak en opname van voedingsstoffen, de activiteit van genen, enz. Een bacterieel onevenwicht - verstoorde darmflora, dysbiose - is gelinkt aan metabole aandoeningen, zoals overgewicht, diabetes, chronische ontsteking en kanker.
Een gezonde, gevarieerde darmflora is ook belangrijk om de bovengenoemde bioactieve stoffen en andere nutriënten uit de voeding om te zetten in nog actievere stoffen."
In het volledige studierapport:
Human microbiomes and their roles in dysbiosis, common diseases, and novel therapeutic approaches
dat gratis is in te zien wordt een mooie analyse gegeven aan de hand van hun bevindingen waarom de darmflora zo essentieel is voor onze gezondheid. En vooral waarom juist de voeding in onze kindertijd en pubertijd cruciaal is in de opbouw van een gezonde darmflora.
Hier het abstract van de studie met de conclusie en een lange referentielijst:
The designing and production of pharmaceuticals based on our own body’s microbiome is an emerging field and is rapidly growing to be fully explored in the near future. This review provides an outlook on recent findings on the human microbiomes, their impact on health and diseases, and on the development of targeted therapies.
Source: Front Microbiol. 2015; 6: 1050. Published online 2015 Oct 6. doi: 10.3389/fmicb.2015.01050
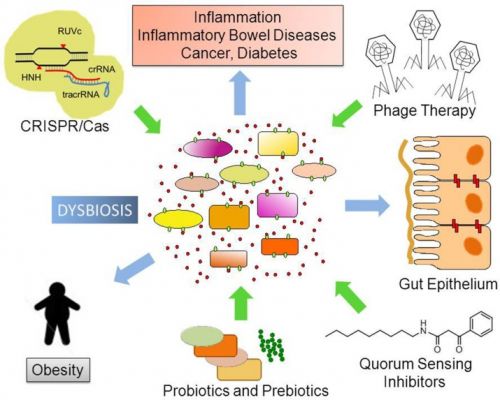
The human body is the residence of a large number of commensal (non-pathogenic) and pathogenic microbial species that have co-evolved with the human genome, adaptive immune system, and diet. With recent advances in DNA-based technologies, we initiated the exploration of bacterial gene functions and their role in human health. The main goal of the human microbiome project is to characterize the abundance, diversity and functionality of the genes present in all microorganisms that permanently live in different sites of the human body. The gut microbiota expresses over 3.3 million bacterial genes, while the human genome expresses only 20 thousand genes. Microbe gene-products exert pivotal functions via the regulation of food digestion and immune system development. Studies are confirming that manipulation of non-pathogenic bacterial strains in the host can stimulate the recovery of the immune response to pathogenic bacteria causing diseases. Different approaches, including the use of nutraceutics (prebiotics and probiotics) as well as phages engineered with CRISPR/Cas systems and quorum sensing systems have been developed as new therapies for controlling dysbiosis (alterations in microbial community) and common diseases (e.g., diabetes and obesity). The designing and production of pharmaceuticals based on our own body’s microbiome is an emerging field and is rapidly growing to be fully explored in the near future. This review provides an outlook on recent findings on the human microbiomes, their impact on health and diseases, and on the development of targeted therapies.
Conclusion and Perspectives
Recent advances in microbiome sequencing projects revealed the high complexity of microbial communities in various human body sites. They have confirmed the critical roles of the human-microbiota ecosystems in health-promoting or disease-causing processes. These studies have highlighted the unexpected and wide-ranging consequences of eliminating certain bacteria living in our body.
While the natural variation of the human microbiota has yet to be fully determined, the annotation and analyses of a large number of human microbiomes have shown that the presence or absence of specific microbial species categorizes human individuals based on enterotypes. It is likely that cultivated and uncultivated microbes will contribute to discovering new fundamental biomarkers for specific human disorders and that they may become better discriminatory tools than human-based ones.
Changes in the stability and dynamic of numerous microbial communities have been associated with several diseases, including type II diabetes, obesity, fatty liver disease, irritable bowel syndrome, and IBDs and even certain cancers. However, further studies need to be done in order to confirm whether low bacterial diversity increases the chances to develop such diseases and metabolic perturbations.
The use of antibiotics compromises genome defense and increases the ability to acquire antibiotic resistance. Prebiotics, probiotics, synbiotics, phage therapy, quorum sensing systems, and CRISPR/Cas systems have been proposed as tools to control and modulate microbial communities. Engineering of pathogen-specific bacteriophages and production of pharmaceuticals based on our own body’s microbiome will be possible and fully explored in the near future. The use of novel pharmaceuticals and nutraceuticals to modulate microbial colonization and development of a healthy gut microbial community in early childhood will support healthy adult human body functions and prevent the occurrence of several diseases.
Author’s Contribution
The authors conducted the literature review process, grading, and categorizing criteria, and quality of selected articles. The authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank colleagues of Institute of Biomedical Sciences of the University of São Paulo for insights and productive discussions. This work was supported by grants from Fundação de Amparo a Pesquisa do Estado de São Paulo (FAPESP, proc. 2015/1177-8, 2015/18647-6) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq).
Footnotes
References
- Aagaard K., Ma J., Antony K. M., Ganu R., Petrosino J., Versalovic J. (2014). The placenta harbors a unique microbiome. Sci. Transl. Med. 6:237ra265 10.1126/scitranslmed.3008599 [PubMed] [Cross Ref]
- Abedon S. T. (2014). Phage therapy: eco-physiological pharmacology. Scientifica (Cairo) 2014:581639 10.1155/2014/581639 [PMC free article] [PubMed] [Cross Ref]
- Abubucker S., Segata N., Goll J., Schubert A. M., Izard J., Cantarel B. L., et al. (2012). Metabolic reconstruction for metagenomic data and its application to the human microbiome. PLoS Comput. Biol. 8:e1002358 10.1371/journal.pcbi.1002358 [PMC free article] [PubMed] [Cross Ref]
- Anukam K. C., Osazuwa E., Osemene G. I., Ehigiagbe F., Bruce A. W., Reid G. (2006). Clinical study comparing probiotic Lactobacillus GR-1 and RC-14 with metronidazole vaginal gel to treat symptomatic bacterial vaginosis. Microbes Infect. 8 2772–2776. 10.1016/j.micinf.2006.08.008 [PubMed] [Cross Ref]
- Arumugam M., Raes J., Pelletier E., Le Paslier D., Yamada T., Mende D. R., et al. (2011). Enterotypes of the human gut microbiome. Nature 473 174–180. 10.1038/nature09944 [PMC free article] [PubMed] [Cross Ref]
- Backhed F., Fraser C. M., Ringel Y., Sanders M. E., Sartor R. B., Sherman P. M., et al. (2012). Defining a healthy human gut microbiome: current concepts, future directions, and clinical applications. Cell Host Microbe 12 611–622. 10.1016/j.chom.2012.10.012 [PubMed] [Cross Ref]
- Belizario J. E. (2013). The humankind genome: from genetic diversity to the origin of human diseases. Genome 56 705–716. 10.1139/gen-2013-0125 [PubMed] [Cross Ref]
- Bi D., Xu Z., Harrison E. M., Tai C., Wei Y., He X., et al. (2012). ICEberg: a web-based resource for integrative and conjugative elements found in Bacteria. Nucleic Acids Res. 40 D621–D626. 10.1093/nar/gkr846 [PMC free article] [PubMed] [Cross Ref]
- Bikard D., Jiang W., Samai P., Hochschild A., Zhang F., Marraffini L. A. (2013). Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res. 41 7429–7437. 10.1093/nar/gkt520 [PMC free article] [PubMed] [Cross Ref]
- Brandt L. J., Reddy S. S. (2011). Fecal microbiota transplantation for recurrent Clostridium difficile infection. J. Clin. Gastroenterol. 45(Suppl.), S159–S167. 10.1097/MCG.0b013e318222e603 [PubMed] [Cross Ref]
- Brown C. T., Sharon I., Thomas B. C., Castelle C. J., Morowitz M. J., Banfield J. F. (2013). Genome resolved analysis of a premature infant gut microbial community reveals a Varibaculum cambriense genome and a shift towards fermentation-based metabolism during the third week of life. Microbiome 1:30 10.1186/2049-2618-1-30 [PMC free article] [PubMed] [Cross Ref]
- Brownawell A. M., Caers W., Gibson G. R., Kendall C. W., Lewis K. D., Ringel Y., et al. (2012). Prebiotics and the health benefits of fiber: current regulatory status, future research, and goals. J. Nutr. 142 962–974. 10.3945/jn.112.158147 [PubMed] [Cross Ref]
- Cani P. D., Delzenne N. M. (2011). The gut microbiome as therapeutic target. Pharmacol. Ther. 130 202–212. 10.1016/j.pharmthera.2011.01.012 [PubMed] [Cross Ref]
- Cani P. D., Neyrinck A. M., Fava F., Knauf C., Burcelin R. G., Tuohy K. M., et al. (2007). Selective increases of bifidobacteria in gut microflora improve high-fat-diet-induced diabetes in mice through a mechanism associated with endotoxaemia. Diabetologia 50 2374–2383. 10.1007/s00125-007-0791-0 [PubMed] [Cross Ref]
- Carr R., Shen-Orr S. S., Borenstein E. (2013). Reconstructing the genomic content of microbiome taxa through shotgun metagenomic deconvolution. PLoS Comput. Biol. 9:e1003292 10.1371/journal.pcbi.1003292 [PMC free article] [PubMed] [Cross Ref]
- Chan B. K., Abedon S. T., Loc-Carrillo C. (2013). Phage cocktails and the future of phage therapy. Future Microbiol 8 769–783. 10.2217/fmb.13.47 [PubMed] [Cross Ref]
- Christen B., Abeliuk E., Collier J. M., Kalogeraki V. S., Passarelli B., Coller J. A., et al. (2011). The essential genome of a bacterium. Mol. Syst. Biol. 7:528 10.1038/msb.2011.58 [PMC free article] [PubMed] [Cross Ref]
- Clemente J. C., Pehrsson E. C., Blaser M. J., Sandhu K., Gao Z., Wang B., et al. (2015). The microbiome of uncontacted Amerindians. Sci. Adv. 1:e1500183 10.1126/sciadv.1500183 [PMC free article] [PubMed] [Cross Ref]
- Clemente J. C., Ursell L. K., Parfrey L. W., Knight R. (2012). The impact of the gut microbiota on human health: an integrative view. Cell 148 1258–1270. 10.1016/j.cell.2012.01.035 [PubMed] [Cross Ref]
- Collison M., Hirt R. P., Wipat A., Nakjang S., Sanseau P., Brown J. R. (2012). Data mining the human gut microbiota for therapeutic targets. Brief. Bioinform. 13 751–768. 10.1093/bib/bbs002 [PubMed] [Cross Ref]
- Comeau A. M., Tetart F., Trojet S. N., Prere M. F., Krisch H. M. (2007). Phage-antibiotic synergy (PAS): beta-lactam and quinolone antibiotics stimulate virulent phage growth. PLoS ONE 2:e799 10.1371/journal.pone.0000799 [PMC free article] [PubMed] [Cross Ref]
- Cucchiara S., Stronati L., Aloi M. (2012). Interactions between intestinal microbiota and innate immune system in pediatric inflammatory bowel disease. J. Clin. Gastroenterol. 46(Suppl.), S64–S66. 10.1097/MCG.0b013e31826a857f [PubMed] [Cross Ref]
- Culligan E. P., Sleator R. D., Marchesi J. R., Hill C. (2014). Metagenomics and novel gene discovery: promise and potential for novel therapeutics. Virulence 5 399–412. 10.4161/viru.27208 [PMC free article] [PubMed] [Cross Ref]
- Curtis T. P., Sloan W. T., Scannell J. W. (2002). Estimating prokaryotic diversity and its limits. Proc. Natl. Acad. Sci. U.S.A. 99 10494–10499. 10.1073/pnas.142680199 [PMC free article] [PubMed] [Cross Ref]
- Dawid S., Roche A. M., Weiser J. N. (2007). The blp bacteriocins of Streptococcus pneumoniae mediate intraspecies competition both in vitro and in vivo. Infect. Immun. 75 443–451. 10.1128/IAI.01775-05 [PMC free article] [PubMed] [Cross Ref]
- De Filippo C., Cavalieri D., Di Paola M., Ramazzotti M., Poullet J. B., Massart S., et al. (2010). Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. U.S.A. 107 14691–14696. 10.1073/pnas.1005963107 [PMC free article] [PubMed] [Cross Ref]
- Defoirdt T., Boon N., Bossier P. (2010). Can bacteria evolve resistance to quorum sensing disruption? PLoS Pathog. 6:e1000989 10.1371/journal.ppat.1000989 [PMC free article] [PubMed] [Cross Ref]
- Delaney M. L., Onderdonk A. B. (2001). Nugent score related to vaginal culture in pregnant women. Obstet. Gynecol. 98 79–84. 10.1016/S0029-7844(01)01402-8 [PubMed] [Cross Ref]
- Dethlefsen L., Huse S., Sogin M. L., Relman D. A. (2008). The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 6:e280 10.1371/journal.pbio.0060280 [PMC free article] [PubMed] [Cross Ref]
- DiGiulio D. B., Romero R., Amogan H. P., Kusanovic J. P., Bik E. M., Gotsch F., et al. (2008). Microbial prevalence, diversity and abundance in amniotic fluid during preterm labor: a molecular and culture-based investigation. PLoS ONE 3:e3056 10.1371/journal.pone.0003056 [PMC free article] [PubMed] [Cross Ref]
- Dominguez-Bello M. G., Costello E. K., Contreras M., Magris M., Hidalgo G., Fierer N., et al. (2010). Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. U.S.A. 107 11971–11975. 10.1073/pnas.1002601107 [PMC free article] [PubMed] [Cross Ref]
- Dong Y. H., Zhang L. H. (2005). Quorum sensing and quorum-quenching enzymes. J. Microbiol. 43 101–109. [PubMed]
- Eckburg P. B., Bik E. M., Bernstein C. N., Purdom E., Dethlefsen L., Sargent M., et al. (2005). Diversity of the human intestinal microbial flora. Science 308 1635–1638. 10.1126/science.1110591 [PMC free article] [PubMed] [Cross Ref]
- Etzold S., Kober O. I., Mackenzie D. A., Tailford L. E., Gunning A. P., Walshaw J., et al. (2014). Structural basis for adaptation of lactobacilli to gastrointestinal mucus. Environ. Microbiol. 16 888–903. 10.1111/1462-2920.12377 [PubMed] [Cross Ref]
- Fardini Y., Chung P., Dumm R., Joshi N., Han Y. W. (2010). Transmission of diverse oral bacteria to murine placenta: evidence for the oral microbiome as a potential source of intrauterine infection. Infect. Immun. 78 1789–1796. 10.1128/IAI.01395-09 [PMC free article] [PubMed] [Cross Ref]
- Fettweis J. M., Serrano M. G., Sheth N. U., Mayer C. M., Glascock A. L., Brooks J. P., et al. (2012). Species-level classification of the vaginal microbiome. BMC Genomics 13(Suppl. 8):S17 10.1186/1471-2164-13-S8-S17 [PMC free article] [PubMed] [Cross Ref]
- Finch R. G., Pritchard D. I., Bycroft B. W., Williams P., Stewart G. S. (1998). Quorum sensing: a novel target for anti-infective therapy. J. Antimicrob. Chemother. 42 569–571. 10.1093/jac/42.5.569 [PubMed] [Cross Ref]
- Flintoft L. (2012). Disease genomics: associations go metagenome-wide. Nat. Rev. Genet. 13 756–757. 10.1038/nrg3347 [PubMed] [Cross Ref]
- Fodor A. A., DeSantis T. Z., Wylie K. M., Badger J. H., Ye Y., Hepburn T., et al. (2012). The “most wanted” taxa from the human microbiome for whole genome sequencing. PLoS ONE 7:e41294 10.1371/journal.pone.0041294 [PMC free article] [PubMed] [Cross Ref]
- Forsberg K. J., Patel S., Gibson M. K., Lauber C. L., Knight R., Fierer N., et al. (2014). Bacterial phylogeny structures soil resistomes across habitats. Nature 509 612–616. 10.1038/nature13377 [PMC free article] [PubMed] [Cross Ref]
- Forslund K., Sunagawa S., Kultima J. R., Mende D. R., Arumugam M., Typas A., et al. (2013). Country-specific antibiotic use practices impact the human gut resistome. Genome Res. 23 1163–1169. 10.1101/gr.155465.113 [PMC free article] [PubMed] [Cross Ref]
- Frantz A. L., Rogier E. W., Weber C. R., Shen L., Cohen D. A., Fenton L. A., et al. (2012). Targeted deletion of MyD88 in intestinal epithelial cells results in compromised antibacterial immunity associated with downregulation of polymeric immunoglobulin receptor, mucin-2, and antibacterial peptides. Mucosal Immunol. 5 501–512. 10.1038/mi.2012.23 [PMC free article] [PubMed] [Cross Ref]
- Fritz J. V., Desai M. S., Shah P., Schneider J. G., Wilmes P. (2013). From meta-omics to causality: experimental models for human microbiome research. Microbiome 1:14 10.1186/2049-2618-1-14 [PMC free article] [PubMed] [Cross Ref]
- Gareau M. G., Sherman P. M., Walker W. A. (2010). Probiotics and the gut microbiota in intestinal health and disease. Nat. Rev. Gastroenterol. Hepatol. 7 503–514. 10.1038/nrgastro.2010.117 [PubMed] [Cross Ref]
- Garneau J. E., Dupuis M. E., Villion M., Romero D. A., Barrangou R., Boyaval P., et al. (2010). The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468 67–71. 10.1038/nature09523 [PubMed] [Cross Ref]
- Gevers D., Pop M., Schloss P. D., Huttenhower C. (2012). Bioinformatics for the human microbiome project. PLoS Comput. Biol. 8:e1002779 10.1371/journal.pcbi.1002779 [PMC free article] [PubMed] [Cross Ref]
- Gomaa A. A., Klumpe H. E., Luo M. L., Selle K., Barrangou R., Beisel C. L. (2014). Programmable removal of bacterial strains by use of genome-targeting CRISPR-Cas systems. MBio 5:e928-13 10.1128/mBio.00928-13 [PMC free article] [PubMed] [Cross Ref]
- Gough E., Shaikh H., Manges A. R. (2011). Systematic review of intestinal microbiota transplantation (fecal bacteriotherapy) for recurrent Clostridium difficile infection. Clin. Infect. Dis. 53 994–1002. 10.1093/cid/cir632 [PubMed] [Cross Ref]
- Grice E. A., Kong H. H., Conlan S., Deming C. B., Davis J., Young A. C., et al. (2009). Topographical and temporal diversity of the human skin microbiome. Science 324 1190–1192. 10.1126/science.1171700 [PMC free article] [PubMed] [Cross Ref]
- Groer M. W., Luciano A. A., Dishaw L. J., Ashmeade T. L., Miller E., Gilbert J. A. (2014). Development of the preterm infant gut microbiome: a research priority. Microbiome 2:38 10.1186/2049-2618-2-38 [PMC free article] [PubMed] [Cross Ref]
- Guani-Guerra E., Santos-Mendoza T., Lugo-Reyes S. O., Teran L. M. (2010). Antimicrobial peptides: general overview and clinical implications in human health and disease. Clin. Immunol. 135 1–11. 10.1016/j.clim.2009.12.004 [PubMed] [Cross Ref]
- Guaraldi F., Salvatori G. (2012). Effect of breast and formula feeding on gut microbiota shaping in newborns. Front. Cell Infect. Microbiol. 2:94 10.3389/fcimb.2012.00094 [PMC free article] [PubMed] [Cross Ref]
- Guo F., Ju F., Cai L., Zhang T. (2013). Taxonomic precision of different hypervariable regions of 16S rRNA gene and annotation methods for functional bacterial groups in biological wastewater treatment. PLoS ONE 8:e76185 10.1371/journal.pone.0076185 [PMC free article] [PubMed] [Cross Ref]
- Haiser H. J., Turnbaugh P. J. (2012). Is it time for a metagenomic basis of therapeutics? Science 336 1253–1255. 10.1126/science.1224396 [PubMed] [Cross Ref]
- Hense B. A., Schuster M. (2015). Core principles of bacterial autoinducer systems. Microbiol. Mol. Biol. Rev. 79 153–169. 10.1128/MMBR.00024-14 [PMC free article] [PubMed] [Cross Ref]
- Hooper L. V., Littman D. R., Macpherson A. J. (2012). Interactions between the microbiota and the immune system. Science 336 1268–1273. 10.1126/science.1223490 [PMC free article] [PubMed] [Cross Ref]
- Human Microbiome Jumpstart Reference Strains C., Nelson K. E., Weinstock G. M., Highlander S. K., Worley K. C., Creasy H. H., et al. (2010). A catalog of reference genomes from the human microbiome. Science 328 994–999. 10.1126/science.1183605 [PMC free article] [PubMed] [Cross Ref]
- Human Microbiome Project C. (2012a). A framework for human microbiome research. Nature 486 215–221. 10.1038/nature11209 [PMC free article] [PubMed] [Cross Ref]
- Human Microbiome Project C. (2012b). Structure, function and diversity of the healthy human microbiome. Nature 486 207–214. 10.1038/nature11234 [PMC free article] [PubMed] [Cross Ref]
- Ishikawa H., Akedo I., Otani T., Suzuki T., Nakamura T., Takeyama I., et al. (2005). Randomized trial of dietary fiber and Lactobacillus casei administration for prevention of colorectal tumors. Int. J. Cancer 116 762–767. 10.1002/ijc.21115 [PubMed] [Cross Ref]
- Jiang W., Bikard D., Cox D., Zhang F., Marraffini L. A. (2013). RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat. Biotechnol. 31 233–239. 10.1038/nbt.2508 [PMC free article] [PubMed] [Cross Ref]
- Jiang W., Ling Z., Lin X., Chen Y., Zhang J., Yu J., et al. (2014). Pyrosequencing analysis of oral microbiota shifting in various caries states in childhood. Microb. Ecol. 67 962–969. 10.1007/s00248-014-0372-y [PubMed] [Cross Ref]
- Joelsson A., Liu Z., Zhu J. (2006). Genetic and phenotypic diversity of quorum-sensing systems in clinical and environmental isolates of Vibrio cholerae. Infect. Immun. 74 1141–1147. 10.1128/IAI.74.2.1141-1147.2006 [PMC free article] [PubMed] [Cross Ref]
- Johnson C. H., Patterson A. D., Idle J. R., Gonzalez F. J. (2012). Xenobiotic metabolomics: major impact on the metabolome. Annu. Rev. Pharmacol. Toxicol. 52 37–56. 10.1146/annurev-pharmtox-010611-134748 [PubMed] [Cross Ref]
- Jones B. V., Marchesi J. R. (2007). Transposon-aided capture (TRACA) of plasmids resident in the human gut mobile metagenome. Nat. Methods 4 55–61. 10.1038/nmeth964 [PubMed] [Cross Ref]
- Kardos N., Demain A. L. (2011). Penicillin: the medicine with the greatest impact on therapeutic outcomes. Appl. Microbiol. Biotechnol. 92 677–687. 10.1007/s00253-011-3587-6 [PubMed] [Cross Ref]
- Kayumov A. R., Khakimullina E. N., Sharafutdinov I. S., Trizna E. Y., Latypova L. Z., Thi Lien H., et al. (2014). Inhibition of biofilm formation in Bacillus subtilis by new halogenated furanones. J. Antibiot. (Tokyo) 68 297–301. 10.1038/ja.2014.143 [PubMed] [Cross Ref]
- Kenyon C., Colebunders R., Crucitti T. (2013). The global epidemiology of bacterial vaginosis: a systematic review. Am. J. Obstet. Gynecol. 209 505–523. 10.1016/j.ajog.2013.05.006 [PubMed] [Cross Ref]
- Kimura N. (2014). Metagenomic approaches to understanding phylogenetic diversity in quorum sensing. Virulence 5 433–442. 10.4161/viru.27850 [PMC free article] [PubMed] [Cross Ref]
- Kootte R. S., Vrieze A., Holleman F., Dallinga-Thie G. M., Zoetendal E. G., de Vos W. M., et al. (2012). The therapeutic potential of manipulating gut microbiota in obesity and type 2 diabetes mellitus. Diabetes Obes. Metab. 14 112–120. 10.1111/j.1463-1326.2011.01483.x [PubMed] [Cross Ref]
- Koren O., Knights D., Gonzalez A., Waldron L., Segata N., Knight R., et al. (2013). A guide to enterotypes across the human body: meta-analysis of microbial community structures in human microbiome datasets. PLoS Comput. Biol. 9:e1002863 10.1371/journal.pcbi.1002863 [PMC free article] [PubMed] [Cross Ref]
- Koskella B., Meaden S. (2013). Understanding bacteriophage specificity in natural microbial communities. Viruses 5 806–823. 10.3390/v5030806 [PMC free article] [PubMed] [Cross Ref]
- Ladizinski B., McLean R., Lee K. C., Elpern D. J., Eron L. (2014). The human skin microbiome. Int. J. Dermatol. 53 1177–1179. 10.1111/ijd.12609 [PubMed] [Cross Ref]
- Le Chatelier E., Nielsen T., Qin J., Prifti E., Hildebrand F., Falony G., et al. (2013). Richness of human gut microbiome correlates with metabolic markers. Nature 500 541–546. 10.1038/nature12506 [PubMed] [Cross Ref]
- Lepage P., Leclerc M. C., Joossens M., Mondot S., Blottiere H. M., Raes J., et al. (2013). A metagenomic insight into our gut’s microbiome. Gut 62 146–158. 10.1136/gutjnl-2011-301805 [PubMed] [Cross Ref]
- Ley R. E., Backhed F., Turnbaugh P., Lozupone C. A., Knight R. D., Gordon J. I. (2005). Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. U.S.A. 102 11070–11075. 10.1073/pnas.0504978102 [PMC free article] [PubMed] [Cross Ref]
- Ley R. E., Hamady M., Lozupone C., Turnbaugh P. J., Ramey R. R., Bircher J. S., et al. (2008). Evolution of mammals and their gut microbes. Science 320 1647–1651. 10.1126/science.1155725 [PMC free article] [PubMed] [Cross Ref]
- Ley R. E., Turnbaugh P. J., Klein S., Gordon J. I. (2006). Microbial ecology: human gut microbes associated with obesity. Nature 444 1022–1023. 10.1038/4441022a [PubMed] [Cross Ref]
- Ma Y., Zhang L., Huang X. (2014). Genome modification by CRISPR/Cas9. FEBS J. 281 5186–5193. 10.1111/febs.13110 [PubMed] [Cross Ref]
- Maidak B. L., Olsen G. J., Larsen N., Overbeek R., McCaughey M. J., Woese C. R. (1996). The ribosomal database project (RDP). Nucleic Acids Res. 24 82–85. 10.1093/nar/24.1.82 [PMC free article] [PubMed] [Cross Ref]
- Manefield M., Rasmussen T. B., Henzter M., Andersen J. B., Steinberg P., Kjelleberg S., et al. (2002). Halogenated furanones inhibit quorum sensing through accelerated LuxR turnover. Microbiology 148(Pt 4), 1119–1127. 10.1099/00221287-148-4-1119 [PubMed] [Cross Ref]
- Manichanh C., Rigottier-Gois L., Bonnaud E., Gloux K., Pelletier E., Frangeul L., et al. (2006). Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut 55 205–211. 10.1136/gut.2005.073817 [PMC free article] [PubMed] [Cross Ref]
- Manor O., Borenstein E. (2015). MUSiCC: a marker genes based framework for metagenomic normalization and accurate profiling of gene abundances in the microbiome. Genome Biol. 16:53 10.1186/s13059-015-0610-8 [PMC free article] [PubMed] [Cross Ref]
- Marchesi J. R., Sato T., Weightman A. J., Martin T. A., Fry J. C., Hiom S. J., et al. (1998). Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl Environ. Microbiol. 64 795–799. [PMC free article] [PubMed]
- Marraffini L. A., Sontheimer E. J. (2008). CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322 1843–1845. 10.1126/science.1165771 [PMC free article] [PubMed] [Cross Ref]
- Mason M. R., Nagaraja H. N., Camerlengo T., Joshi V., Kumar P. S. (2013). Deep sequencing identifies ethnicity-specific bacterial signatures in the oral microbiome. PLoS ONE 8:e77287 10.1371/journal.pone.0077287 [PMC free article] [PubMed] [Cross Ref]
- Meijer K., de Vos P., Priebe M. G. (2010). Butyrate and other short-chain fatty acids as modulators of immunity: what relevance for health? Curr. Opin. Clin. Nutr. Metab. Care 13 715–721. 10.1097/MCO.0b013e32833eebe5 [PubMed] [Cross Ref]
- Mekkes M. C., Weenen T. C., Brummer R. J., Claassen E. (2014). The development of probiotic treatment in obesity: a review. Benef. Microbes 5 19–28. 10.3920/BM2012.0069 [PubMed] [Cross Ref]
- Miller M. B., Bassler B. L. (2001). Quorum sensing in bacteria. Annu. Rev. Microbiol. 55 165–199. 10.1146/annurev.micro.55.1.165 [PubMed] [Cross Ref]
- Mobegi F. M., van Hijum S. A., Burghout P., Bootsma H. J., de Vries S. P., van der Gaast-de Jongh C. E., et al. (2014). From microbial gene essentiality to novel antimicrobial drug targets. BMC Genomics 15:958 10.1186/1471-2164-15-958 [PMC free article] [PubMed] [Cross Ref]
- Montassier E., Batard E., Massart S., Gastinne T., Carton T., Caillon J., et al. (2014). 16S rRNA gene pyrosequencing reveals shift in patient faecal microbiota during high-dose chemotherapy as conditioning regimen for bone marrow transplantation. Microb. Ecol. 67 690–699. 10.1007/s00248-013-0355-4 [PubMed] [Cross Ref]
- Mullany P. (2014). Functional metagenomics for the investigation of antibiotic resistance. Virulence 5 443–447. 10.4161/viru.28196 [PMC free article] [PubMed] [Cross Ref]
- Nagata S., Asahara T., Ohta T., Yamada T., Kondo S., Bian L., et al. (2011). Effect of the continuous intake of probiotic-fermented milk containing Lactobacillus casei strain Shirota on fever in a mass outbreak of norovirus gastroenteritis and the faecal microflora in a health service facility for the aged. Br. J. Nutr. 106 549–556. 10.1017/S000711451100064X [PubMed] [Cross Ref]
- Naik S., Bouladoux N., Wilhelm C., Molloy M. J., Salcedo R., Kastenmuller W., et al. (2012). Compartmentalized control of skin immunity by resident commensals. Science 337 1115–1119. 10.1126/science.1225152 [PMC free article] [PubMed] [Cross Ref]
- Nealson K. H., Hastings J. W. (1979). Bacterial bioluminescence: its control and ecological significance. Microbiol. Rev. 43 496–518. [PMC free article] [PubMed]
- Ng S. C., Hart A. L., Kamm M. A., Stagg A. J., Knight S. C. (2009). Mechanisms of action of probiotics: recent advances. Inflamm. Bowel Dis. 15 300–310. 10.1002/ibd.20602 [PubMed] [Cross Ref]
- Ng W. L., Bassler B. L. (2009). Bacterial quorum-sensing network architectures. Annu. Rev. Genet. 43 197–222. 10.1146/annurev-genet-102108-134304 [PMC free article] [PubMed] [Cross Ref]
- Ochsner U. A., Sun X., Jarvis T., Critchley I., Janjic N. (2007). Aminoacyl-tRNA synthetases: essential and still promising targets for new anti-infective agents. Expert Opin. Investig. Drugs 16 573–593. 10.1517/13543784.16.5.573 [PubMed] [Cross Ref]
- Ostaff M. J., Stange E. F., Wehkamp J. (2013). Antimicrobial peptides and gut microbiota in homeostasis and pathology. EMBO Mol. Med. 5 1465–1483. 10.1002/emmm.201201773 [PMC free article] [PubMed] [Cross Ref]
- Palmer D. J., Metcalfe J., Prescott S. L. (2012). Preventing disease in the 21st century: the importance of maternal and early infant diet and nutrition. J. Allergy Clin. Immunol. 130 733–734. 10.1016/j.jaci.2012.06.038 [PubMed] [Cross Ref]
- Palmer K. L., Gilmore M. S. (2010). Multidrug-resistant enterococci lack CRISPR-cas. mBio 1:e00227-10 10.1128/mBio.00227-10 [PMC free article] [PubMed] [Cross Ref]
- Pandey V., Berwal V., Solanki N., Malik N. S. (2015). Probiotics: healthy bugs and nourishing elements of diet. J. Int. Soc. Prev. Community Dent. 5 81–87. 10.4103/2231-0762.155726 [PMC free article] [PubMed] [Cross Ref]
- Papadimitriou K., Zoumpopoulou G., Foligne B., Alexandraki V., Kazou M., Pot B., et al. (2015). Discovering probiotic microorganisms: in vitro, in vivo, genetic and omics approaches. Front. Microbiol. 6:58 10.3389/fmicb.2015.00058 [PMC free article] [PubMed] [Cross Ref]
- Paredes-Sabja D., Shen A., Sorg J. A. (2014). Clostridium difficile spore biology: sporulation, germination, and spore structural proteins. Trends Microbiol. 22 406–416. 10.1016/j.tim.2014.04.003 [PMC free article] [PubMed] [Cross Ref]
- Parsons J. B., Broussard T. C., Bose J. L., Rosch J. W., Jackson P., Subramanian C., et al. (2014). Identification of a two-component fatty acid kinase responsible for host fatty acid incorporation by Staphylococcus aureus. Proc. Natl. Acad. Sci. U.S.A. 111 10532–10537. 10.1073/pnas.1408797111 [PMC free article] [PubMed] [Cross Ref]
- Perez-Chaparro P. J., Goncalves C., Figueiredo L. C., Faveri M., Lobao E., Tamashiro N., et al. (2014). Newly identified pathogens associated with periodontitis: a systematic review. J. Dent. Res. 93 846–858. 10.1177/0022034514542468 [PMC free article] [PubMed] [Cross Ref]
- Plagens A., Richter H., Charpentier E., Randau L. (2015). DNA and RNA interference mechanisms by CRISPR-Cas surveillance complexes. FEMS Microbiol. Rev. 3 442–463. 10.1093/femsre/fuv019 [PubMed] [Cross Ref]
- Pruesse E., Quast C., Knittel K., Fuchs B. M., Ludwig W., Peplies J., et al. (2007). SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35 7188–7196. 10.1093/nar/gkm864 [PMC free article] [PubMed] [Cross Ref]
- Qi L. S., Larson M. H., Gilbert L. A., Doudna J. A., Weissman J. S., Arkin A. P., et al. (2013). Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152 1173–1183. 10.1016/j.cell.2013.02.022 [PMC free article] [PubMed] [Cross Ref]
- Qin J., Li R., Raes J., Arumugam M., Burgdorf K. S., Manichanh C., et al. (2010). A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464 59–65. 10.1038/nature08821 [PMC free article] [PubMed] [Cross Ref]
- Rahman S. Z., Khan R. A., Gupta V., Uddin M. (2007). Pharmacoenvironmentology–a component of pharmacovigilance. Environ. Health 6:20 10.1186/1476-069X-6-20 [PMC free article] [PubMed] [Cross Ref]
- Rasko D. A., Moreira C. G., Li de R., Reading N. C., Ritchie J. M., Waldor M. K., et al. (2008). Targeting QseC signaling and virulence for antibiotic development. Science 321 1078–1080. 10.1126/science.1160354 [PMC free article] [PubMed] [Cross Ref]
- Ravel J., Gajer P., Abdo Z., Schneider G. M., Koenig S. S., McCulle S. L., et al. (2011). Vaginal microbiome of reproductive-age women. Proc. Natl. Acad. Sci. U.S.A. 108(Suppl. 1), 4680–4687. 10.1073/pnas.1002611107 [PMC free article] [PubMed] [Cross Ref]
- Reddy T. B., Thomas A. D., Stamatis D., Bertsch J., Isbandi M., Jansson J., et al. (2015). The genomes OnLine Database (GOLD) v.5: a metadata management system based on a four level (meta)genome project classification. Nucleic Acids Res. 43 D1099–D1106. 10.1093/nar/gku950 [PMC free article] [PubMed] [Cross Ref]
- Roberfroid M. (2007). Prebiotics: the concept revisited. J. Nutr. 137(3 Suppl. 2), 830S–837S. [PubMed]
- Roberfroid M. B. (2000). Prebiotics and probiotics: are they functional foods? Am. J. Clin. Nutr. 71(6 Suppl.), 1682S–1687S. [PubMed]
- Robinson C. J., Bohannan B. J., Young V. B. (2010). From structure to function: the ecology of host-associated microbial communities. Microbiol. Mol. Biol. Rev. 74 453–476. 10.1128/MMBR.00014-10 [PMC free article] [PubMed] [Cross Ref]
- Romero R., Hassan S. S., Gajer P., Tarca A. L., Fadrosh D. W., Nikita L., et al. (2014). The composition and stability of the vaginal microbiota of normal pregnant women is different from that of non-pregnant women. Microbiome 2:4 10.1186/2049-2618-2-4 [PMC free article] [PubMed] [Cross Ref]
- Rosenthal M., Goldberg D., Aiello A., Larson E., Foxman B. (2011). Skin microbiota: microbial community structure and its potential association with health and disease. Infect. Genet. Evol. 11 839–848. 10.1016/j.meegid.2011.03.022 [PMC free article] [PubMed] [Cross Ref]
- Rouillon C., Zhou M., Zhang J., Politis A., Beilsten-Edmands V., Cannone G., et al. (2013). Structure of the CRISPR interference complex CSM reveals key similarities with cascade. Mol. Cell. 52 124–134. 10.1016/j.molcel.2013.08.020 [PMC free article] [PubMed] [Cross Ref]
- Round J. L., Mazmanian S. K. (2009). The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 9 313–323. 10.1038/nri2515 [PMC free article] [PubMed] [Cross Ref]
- Rupnik M. (2015). Toward a true bacteriotherapy for Clostridium difficile infection. N. Engl. J. Med. 372 1566–1568. 10.1056/NEJMcibr1500270 [PubMed] [Cross Ref]
- Saad R., Rizkallah M. R., Aziz R. K. (2012). Gut Pharmacomicrobiomics: the tip of an iceberg of complex interactions between drugs and gut-associated microbes. Gut Pathog. 4:16 10.1186/1757-4749-4-16 [PMC free article] [PubMed] [Cross Ref]
- Sangiuliano B., Perez N. M., Moreira D. F., Belizario J. E. (2014). Cell death-associated molecular-pattern molecules: inflammatory signaling and control. Mediators Inflamm. 2014:821043 10.1155/2014/821043 [PMC free article] [PubMed] [Cross Ref]
- Sartor R. B., Mazmanian S. K. (2012). Intestinal microbes in inflammatory bowel diseases. Am. J. Gastroenterol. Suppl. 1 15–21. 10.1038/ajgsup.2012.4 [Cross Ref]
- Segata N., Haake S. K., Mannon P., Lemon K. P., Waldron L., Gevers D., et al. (2012). Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biol. 13:R42 10.1186/gb-2012-13-6-r42 [PMC free article] [PubMed] [Cross Ref]
- Selle K., Barrangou R. (2015). Harnessing CRISPR-Cas systems for bacterial genome editing. Trends Microbiol. 23 225–232. 10.1016/j.tim.2015.01.008 [PubMed] [Cross Ref]
- Seo H. S., Xiong Y. Q., Mitchell J., Seepersaud R., Bayer A. S., Sullam P. M. (2010). Bacteriophage lysin mediates the binding of Streptococcus mitis to human platelets through interaction with fibrinogen. PLoS Pathog. 6:e1001047 10.1371/journal.ppat.1001047 [PMC free article] [PubMed] [Cross Ref]
- Slavin J. (2013). Fiber and prebiotics: mechanisms and health benefits. Nutrients 5 1417–1435. 10.3390/nu5041417 [PMC free article] [PubMed] [Cross Ref]
- Smillie C. S., Smith M. B., Friedman J., Cordero O. X., David L. A., Alm E. J. (2011). Ecology drives a global network of gene exchange connecting the human microbiome. Nature 480 241–244. 10.1038/nature10571 [PubMed] [Cross Ref]
- Sokol H., Pigneur B., Watterlot L., Lakhdari O., Bermudez-Humaran L. G., Gratadoux J. J., et al. (2008). Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. U.S.A. 105 16731–16736. 10.1073/pnas.0804812105 [PMC free article] [PubMed] [Cross Ref]
- Sommer M. O., Dantas G. (2011). Antibiotics and the resistant microbiome. Curr. Opin. Microbiol. 14 556–563. 10.1016/j.mib.2011.07.005 [PubMed] [Cross Ref]
- Sulakvelidze A., Alavidze Z., Morris J. G. (2001). Bacteriophage therapy. Antimicrob. Agents Chemother. 45 649–659. 10.1128/AAC.45.3.649-659.2001 [PMC free article] [PubMed] [Cross Ref]
- Thompson A. L., Monteagudo-Mera A., Cadenas M. B., Lampl M. L., Azcarate-Peril M. A. (2015). Milk- and solid-feeding practices and daycare attendance are associated with differences in bacterial diversity, predominant communities, and metabolic and immune function of the infant gut microbiome. Front. Cell Infect. Microbiol. 5:3 10.3389/fcimb.2015.00003 [PMC free article] [PubMed] [Cross Ref]
- Turnbaugh P. J., Hamady M., Yatsunenko T., Cantarel B. L., Duncan A., Ley R. E., et al. (2009). A core gut microbiome in obese and lean twins. Nature 457 480–484. 10.1038/nature07540 [PMC free article] [PubMed] [Cross Ref]
- Turnbaugh P. J., Ley R. E., Hamady M., Fraser-Liggett C. M., Knight R., Gordon J. I. (2007). The human microbiome project. Nature 449 804–810. 10.1038/nature06244 [PMC free article] [PubMed] [Cross Ref]
- van der Oost J., Westra E. R., Jackson R. N., Wiedenheft B. (2014). Unravelling the structural and mechanistic basis of CRISPR-Cas systems. Nat. Rev. Microbiol. 12 479–492. 10.1038/nrmicro3279 [PMC free article] [PubMed] [Cross Ref]
- van Opijnen T., Bodi K. L., Camilli A. (2009). Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat. Methods 6 767–772. 10.1038/nmeth.1377 [PMC free article] [PubMed] [Cross Ref]
- Vercoe R. B., Chang J. T., Dy R. L., Taylor C., Gristwood T., Clulow J. S., et al. (2013). Cytotoxic chromosomal targeting by CRISPR/Cas systems can reshape bacterial genomes and expel or remodel pathogenicity islands. PLoS Genet. 9:e1003454 10.1371/journal.pgen.1003454 [PMC free article] [PubMed] [Cross Ref]
- Verdam F. J., Fuentes S., de Jonge C., Zoetendal E. G., Erbil R., Greve J. W., et al. (2013). Human intestinal microbiota composition is associated with local and systemic inflammation in obesity. Obesity (Silver Spring) 21 E607–E615. 10.1002/oby.20466 [PubMed] [Cross Ref]
- Vuotto C., Longo F., Donelli G. (2014). Probiotics to counteract biofilm-associated infections: promising and conflicting data. Int. J. Oral Sci. 6 189–194. 10.1038/ijos.2014.52 [PubMed] [Cross Ref]
- Wallace B. D., Redinbo M. R. (2013). The human microbiome is a source of therapeutic drug targets. Curr. Opin. Chem. Biol. 17 379–384. 10.1016/j.cbpa.2013.04.011 [PMC free article] [PubMed] [Cross Ref]
- Wang G. (2014). Human antimicrobial peptides and proteins. Pharmaceuticals (Basel) 7 545–594. 10.3390/ph7050545 [PMC free article] [PubMed] [Cross Ref]
- Waters C. M., Bassler B. L. (2005). Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 21 319–346. 10.1146/annurev.cellbio.21.012704.131001 [PubMed] [Cross Ref]
- Whelan K., Quigley E. M. (2013). Probiotics in the management of irritable bowel syndrome and inflammatory bowel disease. Curr. Opin. Gastroenterol. 29 184–189. 10.1097/MOG.0b013e32835d7bba [PubMed] [Cross Ref]
- Wikoff W. R., Anfora A. T., Liu J., Schultz P. G., Lesley S. A., Peters E. C., et al. (2009). Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc. Natl. Acad. Sci. U.S.A. 106 3698–3703. 10.1073/pnas.0812874106 [PMC free article] [PubMed] [Cross Ref]
- Wilson I. D., Nicholson J. K. (2009). The role of gut microbiota in drug response. Curr. Pharm. Des. 15 1519–1523. 10.2174/138161209788168173 [PubMed] [Cross Ref]
- Woese C. R. (1987). Bacterial evolution. Microbiol. Rev. 51 221–271. [PMC free article] [PubMed]
- Woese C. R., Kandler O., Wheelis M. L. (1990). Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl. Acad. Sci. U.S.A. 87 4576–4579. 10.1073/pnas.87.12.4576 [PMC free article] [PubMed] [Cross Ref]
- Wooley J. C., Godzik A., Friedberg I. (2010). A primer on metagenomics. PLoS Comput. Biol. 6:e1000667 10.1371/journal.pcbi.1000667 [PMC free article] [PubMed] [Cross Ref]
- Wozniak R. A., Waldor M. K. (2010). Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat. Rev. Microbiol. 8 552–563. 10.1038/nrmicro2382 [PubMed] [Cross Ref]
- Wright G. D. (2010). Antibiotic resistance in the environment: a link to the clinic? Curr. Opin. Microbiol. 13 589–594. 10.1016/j.mib.2010.08.005 [PubMed] [Cross Ref]
- Xavier K. B., Bassler B. L. (2003). LuxS quorum sensing: more than just a numbers game. Curr. Opin. Microbiol. 6 191–197. 10.1016/S1369-5274(03)00028-6 [PubMed] [Cross Ref]
- Xiao-Jie L., Hui-Ying X., Zun-Ping K., Jin-Lian C., Li-Juan J. (2015). CRISPR-Cas9: a new and promising player in gene therapy. J. Med. Genet. 52 289–296. 10.1136/jmedgenet-2014-102968 [PubMed] [Cross Ref]
- Yatsunenko T., Rey F. E., Manary M. J., Trehan I., Dominguez-Bello M. G., Contreras M., et al. (2012). Human gut microbiome viewed across age and geography. Nature 486 222–227. 10.1038/nature11053 [PMC free article] [PubMed] [Cross Ref]
- Yosef I., Manor M., Kiro R., Qimron U. (2015). Temperate and lytic bacteriophages programmed to sensitize and kill antibiotic-resistant bacteria. Proc. Natl. Acad. Sci. U.S.A. 112 7267–7272. 10.1073/pnas.1500107112 [PMC free article] [PubMed] [Cross Ref]
- Zaura E., Nicu E. A., Krom B. P., Keijser B. J. (2014). Acquiring and maintaining a normal oral microbiome: current perspective. Front. Cell Infect. Microbiol. 4:85 10.3389/fcimb.2014.00085 [PMC free article] [PubMed] [Cross Ref]
- Zhang Q., Rho M., Tang H., Doak T. G., Ye Y. (2013). CRISPR-Cas systems target a diverse collection of invasive mobile genetic elements in human microbiomes. Genome Biol. 14 R40. 10.1186/gb-2013-14-4-r40 [PMC free article] [PubMed] [Cross Ref]
- Zhou Y., Gao H., Mihindukulasuriya K. A., La Rosa P. S., Wylie K. M., Vishnivetskaya T., et al. (2013). Biogeography of the ecosystems of the healthy human body. Genome Biol. 14:R1 10.1186/gb-2013-14-1-r1 [PMC free article] [PubMed] [Cross Ref]
- Zhou Y., Mihindukulasuriya K. A., Gao H., La Rosa P. S., Wylie K. M., Martin J. C., et al. (2014). Exploration of bacterial community classes in major human habitats. Genome Biol. 15 R66. 10.1186/gb-2014-15-5-r66 [PMC free article] [PubMed] [Cross Ref]
- Zoetendal E. G., Rajilic-Stojanovic M., de Vos W. M. (2008). High-throughput diversity and functionality analysis of the gastrointestinal tract microbiota. Gut 57 1605–1615. 10.1136/gut.2007.133603 [PubMed] [Cross Ref]
- Zomer A., Burghout P., Bootsma H. J., Hermans P. W., van Hijum S. A. (2012). ESSENTIALS: software for rapid analysis of high throughput transposon insertion sequencing data. PLoS ONE 7:e43012 10.1371/journal.pone.0043012 [PMC free article] [PubMed] [Cross Ref]
Gerelateerde artikelen
- Alcohol tast de darmbiotica aan en geeft hoger risico op ziektes. Blijkt uit verschillende studies.
- ALDH4A1, een veel voorkomend enzym - eiwit speelt een verrassende rol in het voorkomen van kanker.
- Algemeen: Aangepast voedingspatroon en meer beweging onder telefonische en schriftelijke begeleiding verbetert significant kwaliteit van leven en functioneren van oudere overlevenden van borstkanker en prostaatkanker met overgewicht en slechte levensstijl
- Algemeen: Wat is er met ons voedsel gebeurd? Een desastreuze ontwikkeling lijkt. ODE publiceerde in 2004 een artikel over afname van voedingstoffen van groenten en fruit dat nog steeds actueel lijkt.
- Bloedverdunners die vitamine K in het lichaam remmen worden gelinkt aan een verhoogd risico voor het ontwikkelen en verergeren van (osteo)artrose.
- Boter vervangen door plantaardige olie om te bakken en frituren geeft 17 procent lager risico op sterven aan kanker en overlijden in het algemeen
- Caffeine: mensen met een dieet met een hoog cafeïnegebruik, hebben een rijkere darmflora, met hogere waarden van mogelijk ontstekingsremmende bacteriën, vergeleken met dieet met weinig cafeïne.
- Darmflora: Alzheimerpatiënten missen goede bacterie in hun darmen, die gezonde mensen wel hebben. Veel vezels en gevarieerd eten zou Alzheimer kunnen voorkomen
- Darmflora beinvloed door voeding en leefstijl speelt cruciale rol in ontstaan van ziektes ook bij ontstaan van kanker en behandelingen daarvan
- Deze 6 wetenschappelijke onderzoeken over fysiologisch actieve (voedings)stoffen, zoals vitamines en andere supplementen en toepassingen ervan in de praktijk zijn genomineerd voor de James Lindprijs 2021.
- Dierlijke vetten, roken en leefstijl zijn de belangrijkste factoren die bepalen of iemand kanker krijgt of niet. Blijkt uit heel groot wereldwijd uitgevoerd onderzoek in 187 landen
- Een nieuw drinkvoedingsproduct, met wei-eiwitten, vrije vertakte aminozuren en ursolzuur, verbetert de loopsnelheid en mitochondrieel functioneren van ondervoede ouderen in vergelijking met een standaard drinkvoedingsproduct.
- Eenzaamheid en te weinig sociale contacten blijken grootste risico op overlijden aan alle oorzaken bij mensen met zwaarlijvigheid en obesitas
- Foliumzuur via voeding geeft significant minder risico op hormoon-receptor-negatieve borstkanker bij vrouwen in leeftijd voor de overgang
- Foliumzuur - vitamine B speelt cruciale rol bij optredende mutaties in genen die gerelateerd zijn aan risico op kanker en specifiek bij borstkanker.
- Groene thee vermindert kans op vormen van spijsverteringskanker met gemiddeld 20 procent bij oudere Chinese vrouwen copy 1
- Insulineresistentie: meer dan diabetes type 2. Schatting is dat zeg 75 procent van de Nederlandse bevolking een vorm van insulineresistentie heeft.
- koffieconsumptie lijkt een beschermende factor te zijn tegen de ziekte van Alzheimer en achteruitgang in cognitieve functies.
- Koffie zou het risico op ontstaan van buikvlieskanker met 18 procent kunnen verminderen bewijst een deelstudie uit de Nurses Health Study
- Maak dringend meer werk van gezondheid als onderdeel van bestaanszekerheid, schrijft de Sociaal Economische Raad (SER) in nieuw beleidsrapport
- Mediterraan dieet voorkomt bij vrouwen die dit gebruiken vanaf middelbare leeftijd chronische ziektes en geeft veel betere gezondheid op 70 jaar en ouder
- Mediteraan voedingspatroon kan sterfte aan kanker met 35 procent verminderen in vergelijking met westers dieet.
- Mineralen: De belangrijkste mineralen die de westerse mens dagelijks nodig heeft op een rijtje gezet.
- Multivitamine met mineralen (MVM) gedurende 3 jaar dagelijks ingenomen verminderde bij ouderen (65+) met 60 procent mentale achteruitgang in vergelijking met placebo
- Nitisinon - Orfadin een geneesmiddel tegen een stofwisselingsziekte blijkt in bloed dodelijk voor muggen die malaria verspreiden
- Onderzoek naar effect van voeding op kanker verhelderend. prof. dr. J.W. Coebergh voorspelt de trends van kanker incidentie en kanker bij ouderen. Wat werkt wel en wat werkt niet.
- Overgewicht komt vaker voor dan ondergewicht wereldwijd blijkt uit groot wereldwijd uitgevoerd onderzoek
- RIVM heeft een nieuwe versie van Nederlands Voedingsstoffenbestand (NEVO) gepubliceerd. Hierin informatie over 2152 voedingsmiddelen, waarvan 181 nieuwe producten.
- Plantaardig dieet en tomaten verminderen kans op krijgen van prostaatkanker
- Rapport de Lancet Countdown van VN-klimaatpanel IPCC stelt dat miljoenen doden worden veroorzaakt door slechte luchtkwaliteit, hittegolven en andere gevolgen van CO2-uitstoot waarvan vele doden hadden voorkomen kunnen worden
- Rood vlees en bewerkt vlees verhoogt kans op blaaskanker met 25 tot 33 procent
- Regelmatig noten eten verlaagt de kans om te overlijden aan een hart- of vaatziekte met 29 procent of aan kanker met 11% in vergelijking met mensen die weinig tot geen noten eten
- Screening en diagnostiek via bevolkingsonderzoeken en scans loopt de spuigaten uit en maakt iedere nederlander bijna ziek, stelt de Raad voor Volksgezondheid en Samenleving in een rapport
- Sterk bewerkte voeding en voedingsproducten veroorzaken meer sterfgevallen aan kanker, vooral bij vrouwen met eierstokkanker was de sterfte hoger blijkt uit bevolkingsonderzoek
- Student en leefstijl zorgt met eigen cursussen dat studenten voeding en medicijnen het vak voeding in de zorg kunnen volgen binnen hun opleidingen.
- Suikerconsumptie in voeding is eerder schadelijk dan gunstig voor de gezondheid, vooral bij cardiometabole aandoeningen en kanker heeft toegevoegde suiker schadelijke gevolgen.
- Urinetest kan iemand persoonlijk een voedingspatroon - dieet aanraden. Een zogezegd gepersonaliseerd dieet blijkt uit nieuwe studie.
- Vegetarisch dieet kan klimaat verbeteren en miljoenen mensenlevens redden en kosten gezondheidszorg zouden kunnen dalen met honderden miljarden
- Vegetarische vezelrijke voeding geeft minder risico op diverticulose, afwijkingen aan de darm, blijkt uit grote bevolkingsstudie
- Vetverdeling op het lichaam speelt grotere rol in risico op bepaalde vormen van kanker dan het BMI. Buikvet geeft grootste risico op baarmoederkanker, slokdarmkanker en leverkanker. Heupvet met groter risico op borstkanker.
- Voeding: naleving van verschillende gezonde eetpatronen wordt geassocieerd met een lager risico op totale en oorzaakspecifieke mortaliteit.
- Welke vitamines en mineralen helpen ter voorkoming, preventie van kanker? Een overzicht van belangrijke studies en recente inzichten
- Westerse verschraalde landbouwgrond voorziet de daarop geteelde groenten en fruit niet meer voldoende van noodzakelijke mineralen en vitaminen. Aldus opzienbarend onderzoeksrapport.
- Zwarte bonen bevatten erg veel anti-oxidanten en zijn belangrijk in uw voedingspatroon voor een betere gezondheid.
- Algemeen: Voeding en voedingstoffen die een preventief effect hebben om kanker: te voorkomen. Een aantal studies en aanbevelingen bij elkaar gezet.



Plaats een reactie ...
Reageer op "Darmflora beinvloed door voeding en leefstijl speelt cruciale rol in ontstaan van ziektes ook bij ontstaan van kanker en behandelingen daarvan"